From Journal of Viral Hepatitis
H.-Y. Rao; D.-G. Sun; D. Jiang; R.-F. Yang; F. Guo; J.-H. Wang; F. Liu; H.-Y. Zhang; H.-H. Zhang; S.-C. Du; Q. Jin; H. Qin; A.-S.-F. Lok; L. Wei
Posted: 03/28/2012; J Viral Hepat. 2012;19(3):173-181. © 2012 Blackwell Publishing
Abstract and Introduction
Abstract
Single nucleotide polymorphisms (SNPs) near the IL28B gene have been shown to be associated with response to treatment for chronic hepatitis C and also with spontaneous clearance of hepatitis C virus (HCV) infection. We analysed the association between IL28B genetic variants and spontaneous clearance of HCV infection in 376 HCV-infected Chinese paid plasma donors. Genotyping of eight SNPs near the IL28B region was performed by the iPLEX system (MassARRAY® SNP Genotyping; Sequenom) in all donors, and sequencing was performed on all 80 donors who cleared HCV and on 160 of 296 donors who did not clear HCV to validate the genotypes. Eighty (21.3%) donors spontaneously cleared HCV. Four SNPs were significantly associated with spontaneous HCV clearance: rs8099917 TT (vs GT), rs8105790 TT (vs CT), rs12980275 AA (vs AG) and rs10853728 CC (vs CG or GG) with OR (95% CI) 15.27 (2.07–112.50), 14.88 (2.02–109.72), 7.92 (1.88–33.32) and 2.32 (1.22–4.42) respectively. No association between the other four IL28B SNPs including rs12979860 and spontaneous HCV clearance was found. Women had a higher rate of spontaneous HCV clearance than men [56/213 (26.3%) vs 24/163 (14.6%), P = 0.007], and this was true even after stratification for IL28B genotypes with OR of 1.9–2.2 among those with favourable genotypes. Our results confirmed that IL28B polymorphism is associated with spontaneous clearance of HCV in Chinese subjects, but the SNPs that predict HCV clearance in Chinese subjects were different from those reported in Caucasians. Women were more likely to clear HCV infection regardless of IL28B genotypes.
Introduction
Hepatitis C virus (HCV) infection is rated by the World Health Organization as a global health problem, based on its prevalence, the high rate (50–85%) of chronicity, the rate of severe complications such as cirrhosis and hepatocellular carcinoma, as well as the high costs of antiviral therapy and liver transplantation.[1–3]
There is a significant difference in response to treatment with pegylated interferon plus ribavirin in patients of different race or ethnicity, with sustained virological response rates of 28%, 34%, 52% and 76% in African American, Hispanic, Caucasian American and Asian patients respectively with genotype 1 HCV infection.[4–7] Likewise, racial differences in the rate of spontaneous viral clearance have been reported.[8,9] These racial differences suggest a genetic influence on HCV outcome.
Ge et al.[10] identified a key role for variation in a region close to the interleukin-28B (IL28B) gene on chromosome 19, which codes for interferon-lambda-3, in predicting response to pegylated interferon plus ribavirin therapy in patients with genotype 1 chronic HCV infection. This finding has been confirmed in other independent cohorts.[11–13] In addition, the IL28B single nucleotide polymorphism (SNP) most strongly associated with treatment response, rs12979860, has also been shown to be significantly associated with spontaneous hepatitis C clearance.[14–16] Tanaka et al.[17] found other SNPs in close proximity to IL28B that were associated with treatment response in Japanese patients, with rs8099917 and rs12980275 having the strongest association. Rauch et al.[12] also reported that rs8099917 was significantly associated with treatment response as well as with spontaneous clearance of HCV infection in Swiss and German patients. An association between rs8099917 and spontaneous HCV clearance was also shown in a study of Spanish patients.[18]
Data on the association between IL28B genetic variants and treatment response or spontaneous viral clearance in Chinese patients with HCV infection are limited. In a study of 2371 persons from different continents, Thomas et al.[14] found that the frequency of the C allele at rs12979860 was roughly 95% among Chinese, 70% among Caucasians and 25–40% among Africans. Ge et al.[10] noted that the linkage between rs12979860 and rs8099917 is high among Caucasians but low among African Americans. These data suggest that the IL28B variants that have the strongest association with spontaneous HCV clearance and response to HCV treatment in different racial/ethnic groups may be different.
Between 1972 and 1990, several outbreaks of non-A, non-B hepatitis linked to plasma donation occurred at plasmapheresis centres in Hebei Province Guan County, China. These donors have been followed for 12–19 years after initial documentation of HCV infection and roughly 20% have cleared HCV infection spontaneously. This cohort provided a unique opportunity to evaluate the role of host genetics in the outcome of HCV infection in a population with fairly homogenous ethnic background. Here, we present data on the association between IL28B genetic variation and spontaneous HCV clearance in these HCV-infected plasma donors.
Patients and Methods
Patient Population
Commercial plasma donation was introduced in China in the 1970s. Prior to each donation, the donors were required to have normal alanine aminotransferase (ALT) and no evidence of hepatitis B virus (HBV) infection based on negative test result for hepatitis B surface antigen (HBsAg). Nevertheless, incidences of non-A and non-B hepatitis continued to occur, and multiple outbreaks among six villages and one town occurred in Hebei Province Guan County between 1972 and 1990. The cause of these outbreaks was believed to be related to the return of red cells to donors through contaminated transfusion lines. Post-transfusion non-A, non-B hepatitis outbreaks also occurred among the plasma recipients; however, this study was focused on the donors.
After hepatitis C testing became available, donors from five villages and one town were tested for hepatitis C antibody (anti-HCV) in 1991, and donors from the sixth village were tested for anti-HCV in 1998. Of the 864 registered paid donors, 792 (91.7%) came back for anti-HCV testing and 450 (56.8%) of these 792 tested positive. Donors who tested positive for anti-HCV were invited to return for follow-up assessment every 5–6 years between 1991 and 2008 and once a year thereafter. At each visit, the donors were evaluated and blood was tested for anti-HCV and ALT. HCV RNA was tested during follow-up visits in 2002, 2009 and 2010. All donors who previously tested positive for anti-HCV were invited to participate in a study of IL28B genetic variation during their follow-up visit in 2010. Blood was drawn for biochemical tests, HCV RNA, HCV genotype and IL28B SNPs during the 2010 visit. HCV clearance was defined as undetectable HCV RNA on at least two follow-up evaluations including the visit in 2010 in subjects who were anti-HCV positive during initial screening in 1991 or 1998. All donors in our study were of Han ancestry.
The Ethical Committee of Human Experimentation in Peking University People's Hospital approved the study, and the study was performed in accordance with the Helsinki Declaration of 1975. The plasma donors were enrolled after providing written informed consent for the follow-up study, and additional consent for genetic testing was provided during the 2010 visit.
Biochemical Tests
Fasting blood samples collected at each follow-up visit were tested for ALT, aspartate aminotransferase (AST), γ-glutamyltransferase (γ-GT), alkaline phosphatase, total bilirubin (TBil), direct bilirubin, total protein and albumin by an automated biochemical analyser 7600 (Hitachi, Tokyo, Japan).
Virology Tests
Tests for anti-HCV were carried out using enzyme immunoassay (Chiron, Emeryville, CA, USA) between 1991 and 2005 and chemiluminescent microparticle immunoassay (Architect; Abbott, Chicago, IL, USA) after 2005. HCV RNA was quantified with real-time fluorescence quantitative assay (PG Biotech Company, Shenzhen, China) that has a lower limit of detection of 500 IU/mL between 2002 and 2008 and with Cobas Ampliprep/Cobas Taqman HCV assay (Roche Molecular Diagnostics, Branchburg, NJ, USA) that has a lower limit of detection of 15 IU/mL after 2008. HCV genotype was determined with restriction fragment length polymorphism.[19] HBsAg (Abbott) and anti-HIV were detected by enzyme immunoassay (BioMérieux, Boxtel, the Netherlands).
IL28B SNPs Genotyping
The genomic region associated with response to hepatitis C treatment identified by Ge et al.[10] and Tanaka et al.[17] contains several highly linked SNPs around the IL28B gene. In our cohort, we selected the following SNPs for genotyping: rs12979860, rs10853728, rs11881222, rs12980275, rs4803219, rs4803223, rs8099917 and rs8105790. DNA from the subjects was extracted from peripheral blood using standard methods. Genotyping was performed by the iPLEX system (MassARRAY® SNP Genotyping; Sequenom, San Diego, CA, USA) for all 376 donors. DNA from the donors was blind coded and tested using a 384 format SpectroCHIP™ microarray (Sequenom). A matrix-assisted laser desorption/ionization time-of-flight mass spectrometer was used for data acquisitions from the SpectroCHIP™. Results were analysed using Sequenoms MassARRAY RT™ software (Sequenom). To validate the results of the iPLEX system, we sequenced the region surrounding the IL28B gene that covered the eight SNPs for all donors who cleared HCV and a random selection of approximately 50% of the donors who did not clear HCV. Samples were amplified, and the amplicons were subjected to direct sequencing using the Big Dyes Termination version 1.1 kit (ABI) and the ABI Prism 3730 genetic analyser (Applied Biosystems, Foster City, CA, USA). Sequence results were analysed using Polyphred software (Applied Biosystems). Results of the iPLEX system and direct sequencing were compared.
Statistical Analysis
Genotype frequencies were obtained by direct counting, and statistical analysis was performed by the chi-square test. Qualitative data were also analysed by the chi-square test and odds ratio (OR) with 95% confidence intervals (95% CI). Median values of quantitative variables were compared using a nonparametric test (Mann–Whitney two-tailed test). P-values <0.05 were considered statistically significant. The statistical software package used was SPSS 13.0 for Windows (SPSS Inc., Chicago, IL, USA).
Results
Rate of Spontaneous HCV Clearance
Of the 450 donors who tested positive for anti-HCV in 1991 or 1998, 36 had died (18 because of liver diseases), and 33 were lost to follow-up by May 2010 (Fig. 1). Five donors with HBV co-infection were excluded. The remaining 376 HCV-infected plasma donors were tested for IL28B SNPs, 163 were men and 213 women, with a mean age of 53.2 ± 8.0 years and a mean BMI of 24.5 ± 3.2 (kg/m2) at the time of the study. None of the donors had received hepatitis C treatment because all of them were farmers and their medical insurance did not cover hepatitis C treatment. All the donors were serum anti-HIV negative. Eighty (21.3%) donors had spontaneous clearance of HCV infection (95% CI: 17.1–25.4). All 80 remained anti-HCV positive in 2010; of these, 70 had sample/cutoff (S/CO) ratio ≥5 and 10 had S/CO ratio <5 on the anti-HCV test in 2010.
(Click on picture to enlarge)
Figure 1. Flow diagram showing the disposition of 864 registered plasma donors.
Spontaneous viral clearance was more likely to occur in women than in men, 26.3% (56/213) vs 14.7% (24/163) (P = 0.007). As the donors were not regularly followed up before 1991 or 1998 and most had no symptoms of acute hepatitis during the outbreaks, the onset of infection and the severity of the acute infection could not be ascertained. There was no correlation between age at the time of study and spontaneous HCV clearance. All the donors had ALT tested at least once during the outbreaks, and 153 (40.7%) had elevated ALT on more than one occasion during the outbreaks. The rate of spontaneous HCV clearance was similar in donors with and without elevated ALT during the outbreaks [39/153 (25.5%) vs 41/223 (18.4%) P = 0.10].
During the 2010 visit, donors who had persistent HCV infection had significantly higher mean ALT, AST and γ-GT and lower platelet count than those who cleared HCV. Of the 296 donors who were still HCV RNA positive, 83.8%, 12.8% and 3.4% had HCV genotype 1b, 2a and 1b/2a infection, respectively.
Frequency of IL28B SNPs
Analysis of the IL28B genotype results of 240 donors (80 with spontaneous HCV clearance and 160 with persistent HCV infection) who were tested by both the iPLEX system and by sequencing showed a concordance rate of 97.8–100% for the eight SNPs studied; therefore, the results of the iPLEX system will be presented in this study.
Of the 376 donors, 180 (48.5%, 95% CI: 43.4–53.6) were homozygous for CC, 189 (50.9%, 95% CI: 45.9–56.0) were heterozygous (CT) and only 2 (0.5%, 95% CI: −0.2–1.3) were homozygous for TT at rs12979860, reflecting a C allele frequency of 74.0% (95% CI: 70.8–77.2). Frequency of the other IL28B SNPs is shown inTable 1. No significant deviations in standard genotype quality control according to the Hardy–Weinberg Equilibrium for six SNPs were noted (rs12979860, rs10853728, rs12980275, rs4803223, rs8099917 and rs8105790), while there were significant deviations for two SNPs (rs11881222 and rs4803219).
Association Between IL28B SNPs and Spontaneous Clearance of HCV Infection
Spontaneous HCV clearance was associated with four of the eight IL28B SNPs tested. The IL28B genetic variants rs8099917 [TT vs GT, OR 15.3 (95% CI: 2.1–112.5), P = 0.002], rs8105790 [TT vs CT, OR 14.9 (95% CI: 2.0–109.8), P = 0.0024], rs12980275 [AA vs AG, OR 7.9 (95% CI: 1.9–33.3), P = 0.0043] and rs10853728 [CC vs CG or GG, OR 2.3 (95% CI: 1.2–4.4), P = 0.0206] were associated with a significantly higher rate of spontaneous clearance of HCV (Table 2). Thus, for rs8099917, the SNP with the strongest association with spontaneous HCV clearance, 24.1% of TT homozygous subjects (77/319; 95% CI: 19.4–28.8) and only 2.0% (1/49, 95% CI: 1.9–6.0) of GT heterozygous subjects had spontaneous clearance of HCV.
The other four IL28B SNPs including rs12979860 were not associated with the spontaneous clearance of HCV infection. For rs12979860, spontaneous HCV clearance was observed in 24% (43/180, 95% CI: 17.7–30.1), 19.1% (36/189, 95% CI: 13.5–24.7) and 50% (1/2, 95% CI: 19.3–119.3) subjects with CC, CT and TT genotypes respectively (P = 0.33).
As women had a higher rate of spontaneous HCV clearance than men, the prevalence of favourable genotypes between men and women was compared, and the results showed no significant difference between men and women for all eight SNPs studied (Table 1). Women were significantly more likely to have spontaneous HCV clearance than men even after stratification for IL28B genotype with odds ratio of 1.9–2.2 among those with favourable genotypes (Table 3). As an example, among those with rs8099917 TT genotype, 29.5% women but only 16.9% men had spontaneous HCV clearance, OR 2.1 (95% CI: 1.2–3.6), P = 0.0093.
Haplotype Analysis
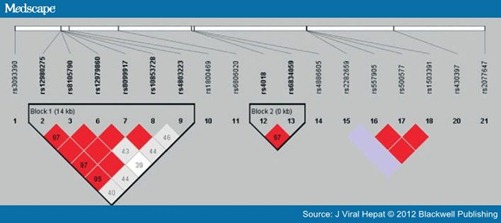
Pair-wise linkage disequilibrium between the SNPs is shown in Fig. 2. Four SNPs (rs12980275, rs8105790, rs12979860 and rs8099917) were in strong linkage disequilibrium. The haplotype ATCT with favourable alleles in these four SNPs was not associated with spontaneous HCV clearance. However, donors with the haplotype ATTC, favourable alleles in the four SNPs that were independently shown to be associated with spontaneous HCV clearance (rs12980275, rs8105790, rs8099917 and rs10853728) were significantly more likely to have spontaneous HCV clearance than those who did not have this haplotype, 24.8% (64/258) vs 13.5% (14/104), OR 2.1 (95% CI: 1.1–4.0), P = 0.018.
Figure 2. Pair-wise linkage disequilibrium pattern of the IL28 region. The degree of linkage disequilibrium between any two SNPs is described by r2 values (among 0.97–1.00). Rs12980275, rs8105790, rs12979860 and rs8099917 are within a genomic block that encompasses the IL28B gene.
Multivariate Analysis of Factors Associated With Spontaneous HCV Clearance
Multivariate analysis including the following variables – gender, the four IL28B SNPs that were significant on univariate analysis and the haplotype ATTC comprising rs12980275, rs8105790, rs8099917 and rs10853728 – showed that IL28B SNP rs8099917 (OR = 14.88, 95% CI: 2.02–109.7, P = 0.008) was the most significant factor associated with spontaneous HCV clearance. Addition of the other three IL28B SNPs or the haplotype ATTC to the SNP rs8099917 did not improve the association with spontaneous HCV clearance.
Gender had an independent effect on spontaneous HCV clearance, the odds ratio for women to clear HCV infection in the multivariate model for all donors was 1.95 (95% CI: 1.14–3.32), for donors with rs8099917 TT was 2.05 (95% CI: 1.18–3.53) and for donors with the haplotype ATTC was 1.97 (95% CI: 1.09–3.55) when compared with men.
Association Between IL28B Genotype and HCV RNA Level and HCV Genotype
Analysis of the HCV RNA levels of the 276 subjects who had detectable HCV RNA at the 2010 visit showed that there was no difference in HCV RNA levels between the subjects with favourable and those with unfavourable genotypes at the four IL28B SNPs shown to be associated with spontaneous HCV clearance. There was also no difference in the distribution of HCV genotypes between the subjects with favourable and those with unfavourable IL28B genotypes.
Discussion
In this study of 376 HCV-infected Chinese paid plasma donors, we found that IL28B gene variants were significantly associated with the spontaneous clearance of HCV infection. The SNP that had the strongest association with spontaneous clearance of HCV infection was rs8099917, and persons with the TT genotype were 15 times more likely to clear HCV than those with the GT genotype. However, the SNP rs12979860 that has been shown to be most strongly associated with response to pegylated interferon and ribavirin and to spontaneous HCV clearance in Caucasians and African Americans was not associated with spontaneous HCV clearance in our study.[10–14]
Thomas et al.[14] studied 1008 subjects in six independent cohorts and found that rs12979860 CC genotype was predictive of spontaneous HCV clearance among individuals of both European and African ancestry. An association between rs12979860 CC genotype and spontaneous HCV clearance was also shown by Tillmann et al.[16] who studied 190 German women and by Montes-Cano et al.[18] in a study of 353 Spanish subjects. In all three studies, only one SNP (rs12979860) was tested. An association between rs8099917 TT genotype and response to pegylated interferon and ribavirin treatment had been reported in the studies of Caucasian patients and Japanese patients with chronic HCV infection.[11,12,17] An association between rs8099917 TT genotype and spontaneous clearance of HCV infection was also demonstrated in a study of 132 predominantly Caucasian patients in Australia and a study of 1362 Swiss and German patients.[12,15] Only two SNPs (rs8099917 and rs12980275) were tested in the study in Australia. The study in Switzerland tested more than 500 000 SNPs. Analysis of data from 1213 individuals with available data at both rs12979860 and rs8099917 found that rs12979860 genotype was highly associated with chronic HCV infection and was in strong linkage disequilibrium with rs8099917.[12]
In this study, we tested eight SNPs and found an association between spontaneous HCV clearance and four SNPs including rs8099917, rs8105790, rs12980275 and rs10853728 but not rs12979860. Ge et al.[10] had noted that the linkage between rs8099917 and rs12979860 is high among Caucasians, but low among African Americans. We found that rs8099917 was strongly linked to rs12979860 (r2 = 1.00), rs12980275 (r2 = 0.97) and rs8105790 (r2 = 1.00) in our study. In our study, all donors with rs12979860 CC had rs8099917 TT genotype, but donors with rs12979860 CT could be rs8099917 TT or TG.
The rs12979860 C allele frequency in the Chinese Han population (83 subjects from Beijing, China) was reported to be 93.4% in HapMap and 94–98% in Thomas' study of 47 Chinese in Taiwan and 59 Chinese in San Francisco, but it was only 74% in our study. The rs8099917 T allele frequency in the Chinese Han population was reported to be 93.5% in HapMap and 93.3% in our study. The subjects in our study all came from one county, Hebei, in the northern part of China. So, the SNP allele frequencies in our study might be only representative of the northern local region. While most Chinese in the major cities of China are of Han ancestry, other ancestries predominate in some parts of China. Studies involving larger numbers of Chinese in different regions of China are necessary to determine the precise frequency of the favourable alleles at rs12979860 and rs8099917 among Chinese of various ancestries.
In this study, the SNP rs8099917 had the strongest association with spontaneous HCV clearance. Although three other SNPs (rs10853728, rs 12980275 and rs8105790) in the IL28B region were independently associated with spontaneous HCV clearance, inclusion of these three SNPs or the haplotype ATTC with favourable alleles at these three SNPs plus rs8099917 did not improve the prediction of spontaneous HCV clearance in the multivariate model compared with rs8099917 alone. Several studies showed that women were more likely to undergo spontaneous HCV clearance than men. One study of 67 persons with acute HCV infection, 66% of whom were injection drug users, found that HCV clearance occurred in 34% of women compared with 3% of men (P < 0.001).[20] Another study followed 135 injection drug users with incident HCV infection and found that the age-adjusted hazard ratio of spontaneous HCV clearance for women was 2.91 (95% CI: 1.68–5.03).[21] A systematic review of 31 longitudinal studies that included 314 subjects showed that the relative risk of spontaneous HCV clearance for men was 0.43 (95% CI: 0.36–0.53) (P = 0.00001).[22]
Two recent studies examined gender, IL28B genetic variants and spontaneous HCV clearance. In Grebely et al.'s[15] study of 132 Australians, rs8099917 TT genotype (adjusted hazard ratio 3.78, 95% CI: 1.04–13.76) was the only factor predicting spontaneous HCV clearance on multivariate analysis, but female gender and jaundice during the acute illness were not. In Montes-Cano et al.'s[18] study of 352 Spanish subjects, rs12979860 CC genotype was associated with a higher rate of spontaneous HCV clearance in both men (72.4%vs 27.6%) and women (72.5%vs 27.5%) compared with those with CT or TT genotype. In this study, we found that women had a higher rate of spontaneous HCV clearance than men, 26.3%vs 14.6%, and this difference persisted even after stratification for IL28B genotypes. Among those with rs8099917 TT genotype, women were twice as likely to have spontaneous HCV clearance as men. Female gender remained an independent predictor of spontaneous HCV clearance in the multivariate model, with OR of 1.95.
Some studies found that the presence of jaundice or symptoms during acute HCV infection was associated with a higher rate of spontaneous HCV clearance.[15,16,22] Tillmann et al.[16] found that jaundice during acute infection was associated with an increased chance of spontaneous HCV clearance (42.9%vs 13.7%) in persons with non-CC genotype at rs12979860, but not in those with CC genotype (56.3%vs 60.6%). In this study, spontaneous HCV clearance rate was similar in donors with and without documented ALT elevation during the outbreaks, but the donors were not regularly monitored, and it is possible that ALT was not tested at the time of acute infection. We were also not able to determine whether any of the donors had symptomatic illness or were jaundiced during the acute infection.
In summary, in this study of a large homogenous cohort of HCV-infected plasma donors, we found that 21.3% had spontaneous HCV clearance. We confirmed that IL28B genotype is associated with spontaneous HCV clearance with rs8099917 TT genotype showing the strongest association, while rs12979860 CC genotype had no association. Women had a higher rate of spontaneous HCV clearance regardless of IL28B genotype. Our results suggest that while IL28B genotype is associated with spontaneous HCV clearance in patients of diverse racial/ethnic background, the SNP with the strongest association may vary. Further studies are needed to confirm this observation and to clarify whether other factors such as gender and severity of the acute illness are independent predictors of spontaneous HCV clearance after correction for IL28B genotype. These results will help in determining which patients with acute HCV infection should start on antiviral therapy and which ones to be monitored.
References
- Ascione A, Tartaglione T, Di Costanzo GG. Natural history of chronic hepatitis C virus infection. Dig Liver Dis 2007; 39(Suppl. 1): S4–S7.
- Sweeting MJ, De Angelis D, Brant LJ, Harris HE, Mann AG, Ramsay ME. The burden of hepatitis C in England. J Viral Hepat 2007; 14: 570–576.
- Global Burden of Hepatitis C Working Group. Global burden of disease for hepatitis C. J Clin Pharmacol 2004; 44: 20–29.
- Conjeevaram HS, Fried MW, Jeffers LJ et al. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology 2006; 131: 470–477.
- Jeffers LJ, Cassidy W, Howell CD, Hu S, Reddy KR. Peginterferon alfa-2a (40 kD) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology 2004; 39: 1702–1708.
- Rodriguez-Torres M, Jeffers LJ, Sheikh MY et al. Peginterferon alfa-2a and ribavirin in Latino and non-Latino whites with hepatitis C. N Engl J Med 2009; 360: 257–267.
- Liu CH, Liu CJ, Lin CL et al. Pegylated interferon-alpha-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: a multicentre, randomized controlled trial. Clin Infect Dis 2008; 47: 1260–1269.
- Thomas DL, Astemborski J, Rai RM et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA 2000; 284: 450–456.
- Aborsangaya KB, Dembinski I, Khatkar S, Alphonse MP, Nickerson P, Rempel JD. Impact of aboriginal ethnicity on HCV core-induced IL-10 synthesis: interaction with IL-10 gene polymorphisms. Hepatology 2007; 45: 623–630.
- Ge D, Fellay J, Thompson AJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 2009; 461: 399–401.
- Suppiah V, Moldovan M, Ahlenstiel G et al. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 2009; 41: 1100–1104.
- Rauch A, Kutalik Z, Descombes P et al. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genomewide association study. Gastroenterology 2010; 138: 1338–1345.
- McCarthy JJ, Li JH, Thompson A et al. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology 2010; 138: 2307–2314.
- Thomas DL, Thio CL, Martin MP et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009; 461: 798–801.
- Grebely J, Petoumenos K, Hellard M et al. Potential role for interleukin- 28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology 2010; 52: 1216–1224.
- Tillmann HL, Thompson AJ, Patel K et al. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology 2010; 139: 1586–1592.
- Tanaka Y, Nishida N, Sugiyama M et al. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 2009; 41: 1105–1109.
- Montes-Cano MA, García-Lozano JR, Abad-Molina C et al. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology 2010; 52: 33–37.
- Du SC, Tao QM, Zhu L. Typing on 5'-terminal noncoding region of hepatitis C virus genome with restrict endonuclease. Zhonghua Yi Xue Za Zhi 1993; 73: 7–9.
- Wang CC, Krantz E, Klarquist J et al. Acute hepatitis C in a contemporary US cohort: modes of acquisition and factors influencing viral clearance. J Infect Dis 2007; 196: 1474–1482.
- Page K, Hahn JA, Evans J et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis 2009; 200: 1216–1226.
- Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat 2006; 13: 34–41.

No comments:
Post a Comment